resubLoss
Resubstitution classification loss for naive Bayes classifier
Description
L= resubLoss(Mdl)L) or the in-sample classification loss, for the naive Bayes classifierMdlusing the training data stored inMdl.Xand the corresponding class labels stored inMdl.Y.
The classification loss (L) is a generalization or resubstitution quality measure. Its interpretation depends on the loss function and weighting scheme; in general, better classifiers yield smaller classification loss values.
Examples
Determine Resubstitution Loss of Naive Bayes Classifier
Determine the in-sample classification error (resubstitution loss) of a naive Bayes classifier. In general, a smaller loss indicates a better classifier.
Load thefisheririsdata set. CreateXas a numeric matrix that contains four petal measurements for 150 irises. CreateYas a cell array of character vectors that contains the corresponding iris species.
loadfisheririsX = meas; Y = species;
Train a naive Bayes classifier using the predictorsXand class labelsY. A recommended practice is to specify the class names.fitcnbassumes that each predictor is conditionally and normally distributed.
Mdl = fitcnb(X,Y,'ClassNames',{'setosa','versicolor','virginica'})
Mdl = ClassificationNaiveBayes ResponseName:‘Y’CategoricalPredictors: [] ClassNames: {'setosa' 'versicolor' 'virginica'} ScoreTransform: 'none' NumObservations: 150 DistributionNames: {'normal' 'normal' 'normal' 'normal'} DistributionParameters: {3x4 cell} Properties, Methods
Mdlis a trainedClassificationNaiveBayesclassifier.
Estimate the in-sample classification error.
L = resubLoss(Mdl)
L = 0.0400
The naive Bayes classifier misclassifies 4% of the training observations.
Determine Resubstitution Logit Loss of Naive Bayes Classifier
Load thefisheririsdata set. CreateXas a numeric matrix that contains four petal measurements for 150 irises. CreateYas a cell array of character vectors that contains the corresponding iris species.
loadfisheririsX = meas; Y = species;
Train a naive Bayes classifier using the predictorsXand class labelsY. A recommended practice is to specify the class names.fitcnbassumes that each predictor is conditionally and normally distributed.
Mdl = fitcnb(X,Y,'ClassNames',{'setosa','versicolor','virginica'});
Mdlis a trainedClassificationNaiveBayesclassifier.
Estimate the logit resubstitution loss.
L = resubLoss(Mdl,'LossFun','logit')
L = 0.3310
The average in-sample logit loss is approximately 0.33.
Input Arguments
Mdl—Full, trained naive Bayes classifier
ClassificationNaiveBayesmodel
Full, trained naive Bayes classifier, specified as aClassificationNaiveBayesmodel trained byfitcnb.
LossFun—Loss function
'classiferror'(default) |'binodeviance'|'exponential'|'hinge'|'logit'|'mincost'|'quadratic'|function handle
Loss function, specified as a built-in loss function name or function handle.
The following table lists the available loss functions. Specify one using its corresponding character vector or string scalar.
Value Description 'binodeviance'Binomial deviance 'classiferror'Classification error 'exponential'Exponential 'hinge'Hinge 'logit'Logistic 'mincost'Minimal expected misclassification cost (for classification scores that are posterior probabilities) 'quadratic'Quadratic 'mincost'is appropriate for classification scores that are posterior probabilities. Naive Bayes models return posterior probabilities as classification scores by default (seepredict).Specify your own function using function handle notation.
Suppose that
nis the number of observations inXandKis the number of distinct classes (numel(Mdl.ClassNames), whereMdlis the input model). Your function must have this signaturewhere:lossvalue =lossfun(C,S,W,Cost)The output argument
lossvalueis a scalar.You specify the function name (
lossfun).Cis ann-by-Klogical matrix with rows indicating the class to which the corresponding observation belongs. The column order corresponds to the class order inMdl.ClassNames.Create
Cby settingC(p,q) = 1如果观察pis in classq,为每一行。设置所有其他元素的行pto0.Sis ann-by-Knumeric matrix of classification scores. The column order corresponds to the class order inMdl.ClassNames.Sis a matrix of classification scores, similar to the output ofpredict.Wis ann-by-1 numeric vector of observation weights. If you passW, the software normalizes the weights to sum to1.Costis aK-by-Knumeric matrix of misclassification costs. For example,Cost = ones(K) - eye(K)specifies a cost of0for correct classification and1for misclassification.
Specify your function using
'LossFun',@.lossfun
For more details on loss functions, seeClassification Loss.
Data Types:char|string|function_handle
More About
Classification Loss
Classification lossfunctions measure the predictive inaccuracy of classification models. When you compare the same type of loss among many models, a lower loss indicates a better predictive model.
Consider the following scenario.
Lis the weighted average classification loss.
nis the sample size.
For binary classification:
yjis the observed class label. The software codes it as –1 or 1, indicating the negative or positive class, respectively.
f(Xj) is the raw classification score for observation (row)jof the predictor dataX.
mj=yjf(Xj) is the classification score for classifying observationjinto the class corresponding toyj. Positive values ofmjindicate correct classification and do not contribute much to the average loss. Negative values ofmjindicate incorrect classification and contribute significantly to the average loss.
For algorithms that support multiclass classification (that is,K≥ 3):
yj*is a vector ofK– 1 zeros, with 1 in the position corresponding to the true, observed classyj. For example, if the true class of the second observation is the third class andK= 4, theny2*= [0 0 1 0]′. The order of the classes corresponds to the order in the
ClassNamesproperty of the input model.f(Xj) is the lengthKvector of class scores for observationjof the predictor dataX. The order of the scores corresponds to the order of the classes in the
ClassNamesproperty of the input model.mj=yj*′f(Xj). Therefore,mjis the scalar classification score that the model predicts for the true, observed class.
The weight for observationjiswj. The software normalizes the observation weights so that they sum to the corresponding prior class probability. The software also normalizes the prior probabilities so they sum to 1. Therefore,
Given this scenario, the following table describes the supported loss functions that you can specify by using the'LossFun'name-value pair argument.
| Loss Function | Value ofLossFun |
Equation |
|---|---|---|
| Binomial deviance | 'binodeviance' |
|
| Exponential loss | 'exponential' |
|
| Classification error | 'classiferror' |
The classification error is the weighted fraction of misclassified observations where is the class label corresponding to the class with the maximal posterior probability.I{x} is the indicator function. |
| Hinge loss | 'hinge' |
|
| Logit loss | 'logit' |
|
| Minimal cost | 'mincost' |
The software computes the weighted minimal cost using this procedure for observationsj= 1,...,n.
The weighted, average, minimum cost loss is
|
| Quadratic loss | 'quadratic' |
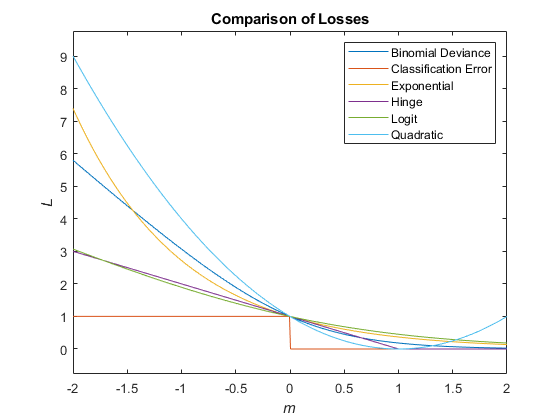
This figure compares the loss functions (except'mincost') for one observation overm. Some functions are normalized to pass through [0,1].
Posterior Probability
Theposterior probabilityis the probability that an observation belongs in a particular class, given the data.
For naive Bayes, the posterior probability that a classification iskfor a given observation (x1,...,xP) is
where:
is the conditional joint density of the predictors given they are in classk.
Mdl.DistributionNamesstores the distribution names of the predictors.π(Y=k) is the class prior probability distribution.
Mdl.Priorstores the prior distribution.is the joint density of the predictors. The classes are discrete, so
Prior Probability
Theprior probabilityof a class is the assumed relative frequency with which observations from that class occur in a population.
See Also
ClassificationNaiveBayes|CompactClassificationNaiveBayes|fitcnb|loss|predict|resubPredict
Topics
Open Example
A modified version of this example exists on your system. Do you want to open this version instead?
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.

Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select:.
Selectweb siteYou can also select a web site from the following list:
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina(Español)
- Canada(English)
- United States(English)
Europe
- Belgium(English)
- Denmark(English)
- Deutschland(Deutsch)
- España(Español)
- Finland(English)
- 弗兰ce(Français)
- Ireland(English)
- Italia(Italiano)
- Luxembourg(English)
- Netherlands(English)
- Norway(English)
- Österreich(Deutsch)
- Portugal(English)
- Sweden(English)
- Switzerland
- United Kingdom(English)
