Improving Classification Trees and Regression Trees
You can tune trees by setting name-value pairs infitctree和fitrtree. The remainder of this section describes how to determine the quality of a tree, how to decide which name-value pairs to set, and how to control the size of a tree.
Examining Resubstitution Error
重新提交错误is the difference between the response training data and the predictions the tree makes of the response based on the input training data. If the resubstitution error is high, you cannot expect the predictions of the tree to be good. However, having low resubstitution error does not guarantee good predictions for new data. Resubstitution error is often an overly optimistic estimate of the predictive error on new data.
Classification Tree Resubstitution Error
This example shows how to examine the resubstitution error of a classification tree.
Load Fisher's iris data.
loadfisheriris
Train a default classification tree using the entire data set.
Mdl = fitctree(meas,species);
检查重新提交错误。
resuberror = resubLoss(Mdl)
resuberror = 0.0200
The tree classifies nearly all the Fisher iris data correctly.
Cross Validation
To get a better sense of the predictive accuracy of your tree for new data, cross validate the tree. By default, cross validation splits the training data into 10 parts at random. It trains 10 new trees, each one on nine parts of the data. It then examines the predictive accuracy of each new tree on the data not included in training that tree. This method gives a good estimate of the predictive accuracy of the resulting tree, since it tests the new trees on new data.
交叉验证了回归树
This example shows how to examine the resubstitution and cross-validation accuracy of a regression tree for predicting mileage based on theCarsmall.数据。
加载Carsmall.data set. Consider acceleration, displacement, horsepower, and weight as predictors of MPG.
loadCarsmall.X = [加速位移马力重量];
使用所有观察来展现回归树。
rtree = fitrtree(x,mpg);
计算样本错误。
resuberror = resubLoss(rtree)
resuberror = 4.7188
回归树的重新提交损失是平均误差。生成的值表示树的典型预测误差是关于4.7的平方根,或者超过2的一点。
Estimate the cross-validation MSE.
RNG.'默认';cvrtree = crossval(rtree);cvloss = kfoldloss(cvrtree)
cvloss = 23.5706.
交叉验证的损失几乎为25,这意味着在新数据上的树的典型预测误差约为5.这表明交叉验证的损失通常高于简单的resubStutitut损失。
Choose Split Predictor Selection Technique
标准的车往往选择contin算法uous predictors that have many levels. Sometimes, such a selection can be spurious and can also mask more important predictors that have fewer levels, such as categorical predictors. That is, the predictor-selection process at each node is biased. Also, standard CART tends to miss the important interactions between pairs of predictors and the response.
为了减轻选择偏差并增加重要交互的检测,您可以使用曲率或交互测试的用法使用'PredictorSelection'name-value pair argument. Using the curvature or interaction test has the added advantage of producing better predictor importance estimates than standard CART.
This table summarizes the supported predictor-selection techniques.
| Technique | 'PredictorSelection'值 |
Description | Training speed | When to specify |
|---|---|---|---|---|
| 标准推车[1] | 默认 | Selects the split predictor that maximizes the split-criterion gain over all possible splits of all predictors. |
Baseline for comparison | Specify if any of these conditions are true:
|
| Curvature test[2][3] | 'curvature' |
Selects the split predictor that minimizes thep-value of chi-square tests of independence between each predictor and the response. | Comparable to standard CART | Specify if any of these conditions are true:
|
| Interaction test[3] | 'interaction-curvature' |
Chooses the split predictor that minimizes thep- 在每个预测器和响应之间的独立性的Chi-Square测试(即,进行曲率测试),并最小化p-value of a chi-square test of independence between each pair of predictors and response. | Slower than standard CART, particularly when data set contains many predictor variables. | Specify if any of these conditions are true:
|
For more details on predictor selection techniques:
For classification trees, see
PredictorSelection和节点拆分规则.对于回归树,看
PredictorSelection和节点拆分规则.
Control Depth or “Leafiness”
When you grow a decision tree, consider its simplicity and predictive power. A deep tree with many leaves is usually highly accurate on the training data. However, the tree is not guaranteed to show a comparable accuracy on an independent test set. A leafy tree tends to overtrain (or overfit), and its test accuracy is often far less than its training (resubstitution) accuracy. In contrast, a shallow tree does not attain high training accuracy. But a shallow tree can be more robust — its training accuracy could be close to that of a representative test set. Also, a shallow tree is easy to interpret. If you do not have enough data for training and test, estimate tree accuracy by cross validation.
fitctree和fitrtreehave three name-value pair arguments that control the depth of resulting decision trees:
MaxNumSplits— The maximal number of branch node splits isMaxNumSplits每棵树。setMaxNumSplitsto get a deep tree. The default issize(X,1) – 1.minleafsize.- 每个叶子至少有minleafsize.观察。设置小值minleafsize.to get deep trees. The default is1.MinParentSize- 树中的每个分支节点至少有MinParentSize观察。设置小值MinParentSizeto get deep trees. The default is10.
如果您指定MinParentSize和minleafsize., the learner uses the setting that yields trees with larger leaves (i.e., shallower trees):
MinParent = max(MinParentSize,2*MinLeafSize)
If you supplyMaxNumSplits, the software splits a tree until one of the three splitting criteria is satisfied.
For an alternative method of controlling the tree depth, seePruning.
选择适当的树深度
This example shows how to control the depth of a decision tree, and how to choose an appropriate depth.
加载ionosphere数据。
loadionosphere
Generate an exponentially spaced set of values from10through100.that represent the minimum number of observations per leaf node.
leafs = logspace(1,2,10);
Create cross-validated classification trees for theionosphere数据。指定使用最小叶子尺寸生长每棵树leafs.
RNG('默认') N = numel(leafs); err = zeros(N,1);forn=1:N t = fitctree(X,Y,'CrossVal','On',...'MinLeafSize',叶子(n));err(n)= kfoldloss(t);endplot(leafs,err); xlabel('Min Leaf Size'); ylabel('cross-validated error');
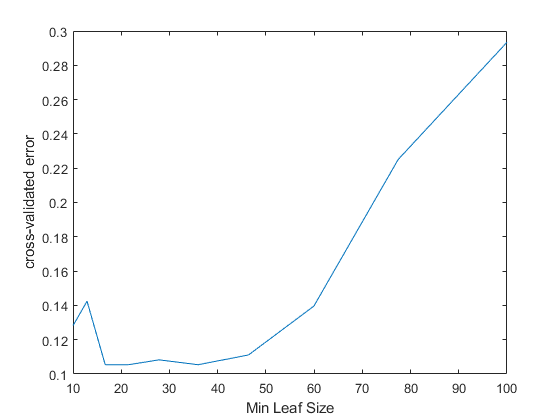
最好的叶子尺寸在约20和50每叶的观察。
至少比较近乎最佳树40observations per leaf with the default tree, which uses10observations per parent node and1每叶观察。
默认Tree = fitctree(X,Y); view(DefaultTree,'Mode','Graph')

OptimalTree = fitctree(X,Y,'MinLeafSize',40); view(OptimalTree,'模式','graph')
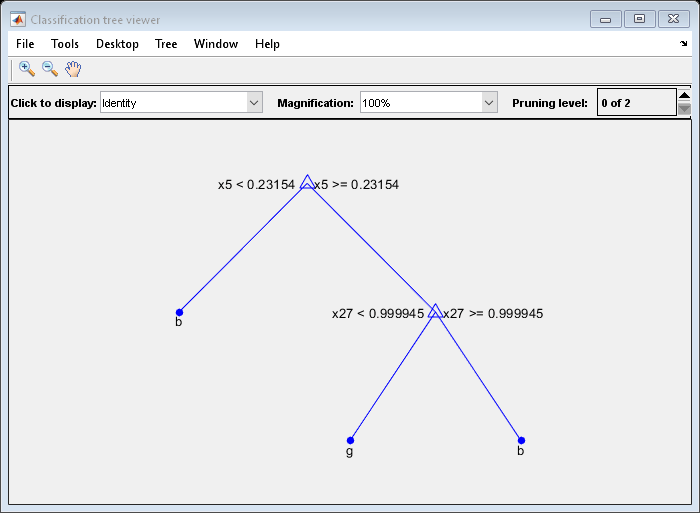
resubOpt = resubLoss(OptimalTree); lossOpt = kfoldLoss(crossval(OptimalTree)); resubDefault = resubLoss(DefaultTree); lossDefault = kfoldLoss(crossval(DefaultTree)); resubOpt,resubDefault,lossOpt,lossDefault
resubOpt = 0.0883
resubDefault = 0.0114
lossOpt = 0.1054
lossDefault = 0.1054
The near-optimal tree is much smaller and gives a much higher resubstitution error. Yet, it gives similar accuracy for cross-validated data.
Pruning
修剪通过在同一树枝上合并叶子来优化树深度(叶粉)。Control Depth or “Leafiness”describes one method for selecting the optimal depth for a tree. Unlike in that section, you do not need to grow a new tree for every node size. Instead, grow a deep tree, and prune it to the level you choose.
使用命令行的修剪树修剪方法(分类)或修剪方法(回归)。或者,与树查看器交互式修剪树:
查看(树,'模式','graph')
要修剪树,树必须包含修剪序列。默认情况下,两者都是fitctree和fitrtree在施工期间计算树的修剪序列。如果你用的树'Prune'名称 - 值对设置为'off', or if you prune a tree to a smaller level, the tree does not contain the full pruning sequence. Generate the full pruning sequence with the修剪方法(分类)或修剪方法(回归)。
Prune a Classification Tree
This example creates a classification tree for theionospheredata, and prunes it to a good level.
加载ionospheredata:
loadionosphere
Construct a default classification tree for the data:
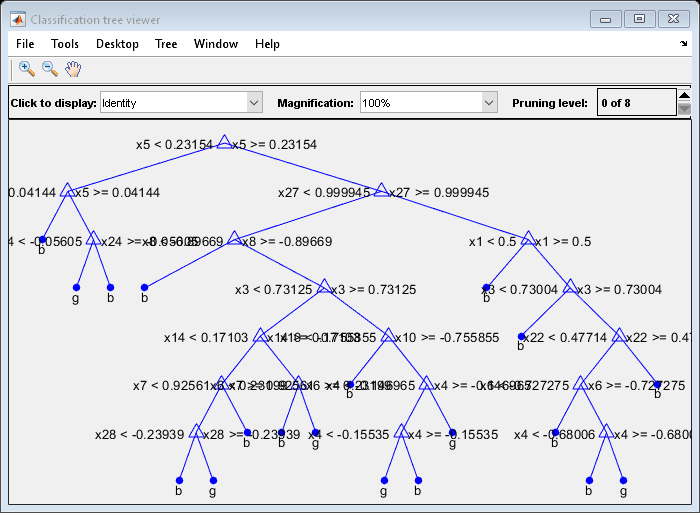
tree = fitctree(X,Y);
View the tree in the interactive viewer:
查看(树,'Mode','Graph')
Find the optimal pruning level by minimizing cross-validated loss:
[~,~,~,bestlevel] = cvLoss(tree,...'SubTrees','All','TreeSize','min')
Bestlevel = 6.
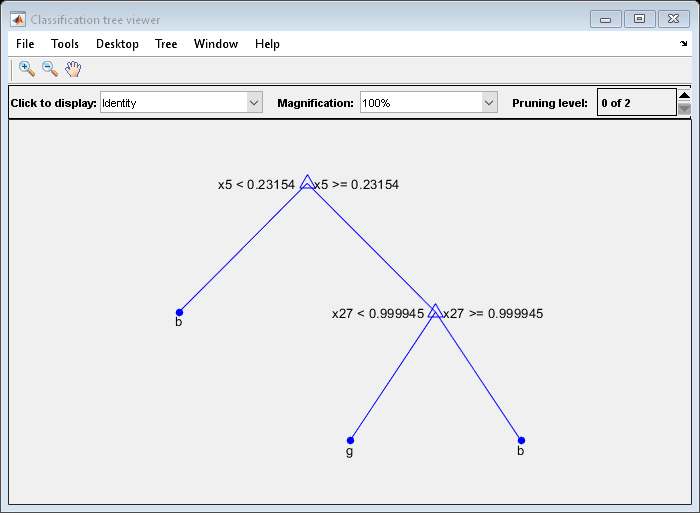
Prune the tree to level6:
查看(树,'Mode','Graph','Prune'6)

Alternatively, use the interactive window to prune the tree.
修剪树算法tr是一样的ee in the "Select Appropriate Tree Depth" example.
Set'TreeSize'to'SE'(默认值)找到树错误从最佳级别不超过错误的最大修剪级别加上一个标准偏差:
[~,~,~,bestlevel] = cvLoss(tree,'SubTrees','All')
Bestlevel = 6.
在这种情况下,级别对于任何一个设置都是相同的'TreeSize'.
Prune the tree to use it for other purposes:
树= prune(树,'Level'6);查看(树,'Mode','Graph')
参考资料
[1]Breiman, L., J. H. Friedman, R. A. Olshen, and C. J. Stone.分类和回归树。Boca Raton, FL: Chapman & Hall, 1984.
[2] LOH,W.Y.和Y.S.Shih。“分类树的分离选择方法。”STATISTICA SINICA., Vol. 7, 1997, pp. 815–840.
[3] LOH,W.Y.“具有无偏的变量选择和相互作用检测的回归树。”STATISTICA SINICA., Vol. 12, 2002, pp. 361–386.
See Also
ClassificationTree.|fitctree|fitrtree|predict (CompactClassificationTree)|预测(CompactregressionTree)|修剪(ClassificationTree)|修剪(RegressionTree)|回归植物

